
Contact information
peijun.zhang@strubi.ox.ac.uk
peijun.zhang@diamond.ac.uk
Professor Peijun Zhang Research
Henry Wellcome Building of Genomic Medicine
Colleges
 |
| Complete all-atom model of HIV-1 capsid (Nature 497:643, 2013, featured on Nature Cover). |
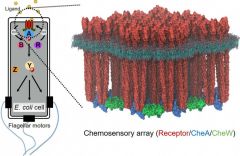 |
| Bacterial chemosensory arrays (Elife 4:e08419, 2015) |
| HIV-1 nuclear import is selective and depends on both capsid elasticity and nuclear pore adaptability (Nat Microbiol. 10(8):1868, 2025) |
Peijun Zhang
Professor of Structural Biology
Structural Biology of Human Pathogens
Our research is aimed at an integrated, atomistic understanding of molecular mechanisms of virus and bacteria infections by developing and combining novel technologies for high-resolution cryoEM and cryo-electron tomography with complementary computational and biophysical/biochemical methods. Current research efforts in our lab are directed to:
HIV-1 capsid assembly, maturation, and interactions with host cell factors
Retroviruses, such as human immunodeficiency virus 1 (HIV-1), contain mature conical capsids that enclose the viral RNA genome, enzymes and accessory proteins. The assembly and stability of the viral capsid are critical to the viral replication life cycle. Structures of the building blocks of the capsid assembly were determined to atomic level, the mechanisms of capsid assembly and disassembly during a productive infection, however, remain unclear. Such information is essential for the development of therapeutic drugs that target viral capsid. More importantly, the surface of HIV-1 capsid serves a primary interaction interface between the virus and the host cell. Many host defense proteins have been identified to interact with the viral capsid and block HIV-1 infection. Yet, very little is known about their precise recognition and interactions, and thus mechanisms of inhibition. We are developing cutting-edge cryoEM technologies that bring unprecedented resolution and enable in situ structures of HIV-1 and in complex with host proteins, such as CypA, TRIM5α, TRIMCyp, CPSF6 and MxB, to decipher their underlining functional roles
Bacterial chemotaxis sensory arrays
Bacteria use chemotaxis signaling pathways to monitor their environment and respond appropriately to change, which is crucial for colonization and infection for bacterial pathogens. The essential core signaling unit comprises transmembrane receptors, a histidine kinase CheA, and a coupling protein CheW. Remarkably, bacteria accomplish the extraordinary gain and cooperativity in chemotaxis signaling by arranging a few hundred core signaling units into higher order arrays localized at the cell pole. We aim to determine the precise molecular mechanisms of chemotaxis cooperative signaling using high-resolution cryoEM and cryoET in combination with site-directed mutagenesis and computational modeling. Our long-term goal is to develop plausible molecular models, at atomic resolution, for the entire signaling pathway by assembling structural “snapshots” of the signaling states.
Methods development for in situ structural biology
Driven by biological questions and inspired by the bottlenecks we have to overcome, we devote significant efforts to the advancement of novel cryoEM methods and technologies, in particulat structural biology in the cellular context. We are working on a wide spectrum of technical advances that will be essential to realize the promise of imaging cells and tissues at molecular resolutions, including correlative microscopy, cryo-FIB/SEM and high resolution sub-tomogram classification and averaging.
Major funding
 Wellcome Discovery Award: Resolving HIV-1 transport and host cofactor regulation in the cellular context
Wellcome Discovery Award: Resolving HIV-1 transport and host cofactor regulation in the cellular context
 NIH/NIAID: University of Pittsburgh Center for HIV Protein Interactions (PCHPI)-CryoEM Core; Correlative cryoET of the HIV-1 integration targeting in native T-lymphocytes
NIH/NIAID: University of Pittsburgh Center for HIV Protein Interactions (PCHPI)-CryoEM Core; Correlative cryoET of the HIV-1 integration targeting in native T-lymphocytes
 ERC AdG Grant: Molecular choreography of bacterial chemotaxis signalling
ERC AdG Grant: Molecular choreography of bacterial chemotaxis signalling
Key publications
-
Structural basis for HIV-1 capsid adaption to a deficiency in IP6 packaging.
Journal article
Zhu Y. et al, (2025), Nat Commun, 16
Recent publications
-
Structural basis for HIV-1 capsid adaption to a deficiency in IP6 packaging.
Journal article
Zhu Y. et al, (2025), Nat Commun, 16
-
Direct visualization of HIV-1 core nuclear import and its interplay with the nuclear pore.
Journal article
Hou Z. et al, (2025), EMBO Rep
-
In-cell structure and variability of pyrenoid Rubisco.
Journal article
Elad N. et al, (2025), Nat Commun, 16
-
Covalently constrained 'Di-Gembodies' enable parallel structure solutions by cryo-EM.
Journal article
Yi G. et al, (2025), Nat Chem Biol
-
HIV-1 nuclear import is selective and depends on both capsid elasticity and nuclear pore adaptability.
Journal article
Hou Z. et al, (2025), Nature microbiology
-
Unveiling the structural spectrum of SARS-CoV-2 fusion by in situ cryo-ET.
Journal article
Akıl C. et al, (2025), Nat Commun, 16
-
In situ cryo-electron microscopy and tomography of cellular and organismal samples.
Journal article
Majumder P. and Zhang P., (2025), Current opinion in structural biology, 93
-
In-cell chromatin structure by Cryo-FIB and Cryo-ET
Journal article
Hou Z. and Zhang P., (2025), Current Opinion in Structural Biology, 92
-
Structural maturation of the matrix lattice is not required for HIV-1 particle infectivity.
Journal article
Chen L. et al, (2025), Sci Adv, 11
-
Bacterial pathogen deploys the iminosugar glycosyrin to manipulate plant glycobiology.
Journal article
Sanguankiattichai N. et al, (2025), Science (New York, N.Y.), 388, 297 - 303
-
Correlative In Situ Cryo-ET Reveals Cellular and Viral Remodeling Associated with Selective HIV-1 Core Nuclear Import.
Preprint
Hou Z. et al, (2025)
-
In-cell Structure and Variability of Pyrenoid Rubisco.
Preprint
Elad N. et al, (2025)
-
Unveiling the Complete Spectrum of SARS-CoV-2 Fusion Stages by In Situ Cryo-ET.
Preprint
Akıl C. et al, (2025)
-
Bacterial pathogen deploys iminosugar galactosyrin to manipulate plant glycobiology
Preprint
Sanguankiattichai N. et al, (2025)
-
Structural basis for HIV-1 capsid adaption to rescue IP6-packaging deficiency.
Preprint
Zhu Y. et al, (2025)
-
Structural maturation of the matrix lattice is not required for HIV-1 particle infectivity.
Preprint
Chen L. et al, (2024)
-
Rubisco packaging and stoichiometric composition of the native β-carboxysome in Synechococcus elongatus PCC7942.
Journal article
Sun Y. et al, (2024), Plant physiology
-
Open architecture of archaea MCM and dsDNA complexes resolved using monodispersed streptavidin affinity CryoEM
Journal article
Ma J. et al, (2024), Nature Communications, 15
-
Adaptive multi-epitope targeting and avidity-enhanced nanobody platform for ultrapotent, durable antiviral therapy.
Journal article
Xiang Y. et al, (2024), Cell
-
Rubisco packaging and stoichiometric composition of a native β-carboxysome.
Preprint
Sun Y. et al, (2024)



